A research team in the University of Macau (UM) Faculty of Health Sciences (FHS), led by Associate Professor Xiaoling XU, has discovered a tumour evolution process through which the tumours initiate from cells with single nucleotide variants (SNVs) by performing simultaneous single nucleotide variant (SNV) and copy number variation (CNV) analysis. The team has also identified a novel tumour metastasis suppressor ‘PLEKHA5’, whose deficiency promotes cancer metastasis to the liver and/or lung. This discovery represents a significant progress in breast cancer metastasis research. The study has been published in the internationally renowned journal Nature Communications.
Breast cancer gene 1 (BRCA1) mutation is the main cause of nearly 25 per cent of hereditary familial breast cancer cases, and the estimated lifetime risk of developing breast cancer for women with BRCA1 germline mutations is between 40 and 80 per cent. The driver events vary from person to person, and even in the same patient, the driver events for different tumours might be different and need to be elucidated. At present, the biology governing the initiation of BRCA1-associated tumours and the evolution from the early premalignant stage to tumour progression and metastasis stage are not fully understood. Over the past few years, single-cell whole-exome sequencing (scWES) has become a powerful approach to deciphering intratumour heterogeneity and identifying cancer drivers. However, simultaneous analysis of single nucleotide variants (SNVs) and copy number variations (CNVs) of a single cell remains challenging.
To understand the genetic features and evolution of BRCA1-deficient breast cancers and identify their drivers, the research team performed simultaneous SNV and CNV analysis in bulk and single cells of BRCA1-deficient mammary glands and tumours using WES; they also conducted functional validation for candidate driver genes with CRISPR-Cas9-mediated knockouts in vitro and in vivo. Several notable findings that have not been clearly illustrated previously have been uncovered in this study. These include: 1) A tumour evolution process was identified through which tumours initiate from cells with SNVs affecting putative driver genes during premalignant stages and malignantly progress later via CNVs acquired in chromosomal regions with many cancer driver genes. 2) These are random events that hit many putative cancer drivers besides p53 to generate unique genetic and pathological features for each tumour. 3) Cancer drivers or metastasis drivers, when they are present at low frequency or occur during premalignant stages, could be missed by bulk DNA sequencing due to high heterogeneity but can be identified by analysing only a small number of single cells. 4) One of the examples is the PLEKHA5 gene, the mutation of which is present in a small population of cells in the primary tumour but became dominant in liver metastases. 5) The scWES results combined with CRISPR-Cas9-mediated knockout studies provide solid evidence that PLEKHA5 does not affect primary tumour growth; however, it is a metastasis suppressor, whose deficiency promotes cancer metastasis to the liver and/or lung.
This study was led by Associate Professor Xiaoling XU, and PhD student Jianlin LIU is the first author. FHS Chair Professor Chuxia DENG, Associate Professor Chris Koon Ho WONG, Research Assistant Professor Kai MIAO, research assistant Ragini ADHAV, and PhD student Sek Man SU also made important contributions to the project. This study was funded by UM (file no: MYRG2016-00138-FHS、MYRG2017-00088-FHS、 MYRG2019-00064-FHS) and the Science and Technology Development Fund (FDCT), Macao SAR (file no: 027/2015/A1, 029/2017/A1, 0101/2018/A3, and 0011/2019/AKP).
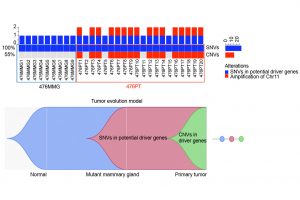
The team has discovered a tumour evolution process by performing simultaneous SNV and CNV analysis
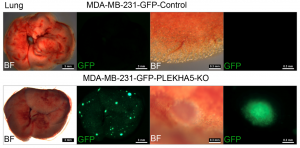
The team has identified a novel tumour metastasis suppressor ‘PLEKHA5’

Xu Xiaoling (2nd row, 2nd from left) and Liu Jianlin (2nd row, 3rd from left)

