A team led by Chuxia DENG, Dean of the Faculty of Health Sciences (FHS) of the University of Macau (UM), has made a significant breakthrough in breast cancer research by uncovering a new mechanism for the occurrence and development of this type of cancer. Based on this finding, the team has discovered a new drug treatment strategy, which brings good news for breast cancer patients. The study has received considerable attention in the field and has been published in the international journal Science Advances.
Breast cancer is the accumulated consequence of malignant and intricate events caused by multiple genetic alterations. Previous studies have found that genes such as BRCA1, BRCA2, p53, CHK2, and 53BP1 are involved in the tumourigenesis process, with BRCA1 being a critical tumour suppressor in breast cancer. Traditional genetic engineering methods detect candidate genes one by one, which is inefficient and time consuming. To compare the contribution of genes under the same biological pressure, Prof Deng’s team conducted an unbiased in vivo screening by integrating the sleeping beauty system (SB+;T2Onc3+) into the Cre-LoxP mediated Brca1 mammary-gland-specific knockout (Brca1Flox11/Flox11; MMTV-Cre and Brca1Flox11/Flox11;Wap-Cre) mouse model. They analysed the 169 genes on the list and found that Cullin-5 (Cul5), which was ranked fourth on the list, acted as a top candidate tumour suppressor, whose disruption enhanced BRCA1-related tumourigenesis. This notion was confirmed by a parallel whole-genome screening mediated by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9.
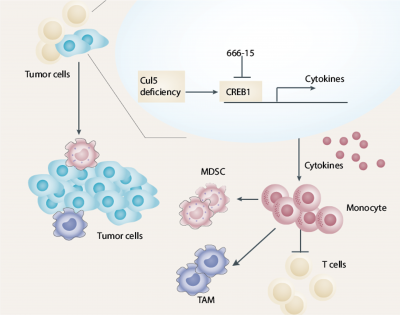
The team also found that Cul5-knockout (KO) cells showed a similar proliferation rate as wild-type (WT) cells, yet Cul5-KO tumours developed much faster than Cul5-WT tumours. These data suggested that the tumour microenvironment (TME) might play a major role in promoting the growth of Cul5-KO tumours. The BALB/c-4T1 and BALB/c-EMT6 mouse models showed that Cul5 deficiency activates CREB1-CCL2 signalling, resulting in the accumulation of monocytes and polymorphonuclear myeloid-derived suppressor cells and the reduction of T cells that lead to cancer occurrence and development. The models also showed that the important downstream factors, CREB1 and CCL2, are involved in the TME process, with Cul5 acting as a ubiquitination-linked enzyme that ubiquitinates CREB1 and degrades CREB1 to inhibit tumour development under regular and stressful conditions. Cul5 deficiency allows the activation of the CREB1-CCL2 pathway, disrupting the balance of TME and leading to tumour growth.
In clinical breast cancer samples, Cul5 usually functions as a tumour suppressor, and its expression often remains at a relatively low level. The activation of CUL5 or inhibition of downstream activities may have applications in the treatment of tumours with low levels of CUL5. Since there is currently no suitable agonist for activating CUL5, the team focused on 666-15, an inhibitor of CREB1, to suppress cancer growth. In summary, this study has revealed the critical role of CUL5 in the occurrence and development of breast cancer. Based on this finding, the research team discovered a new drug treatment strategy for breast cancer through research on tumour immunology and demonstrated that 666-15, the specific inhibitor CREB1, could treat CUL5-deficient breast cancer models, providing early and new ideas for the clinical treatment of breast cancer.
Prof. Deng is the corresponding author of the study, and PhD student Si CHEN is the first author. FHS Associate Professor Xiaoling XU, Assistant Professor Kai MIAO, postdoctoral fellow Fangyuan SHAO, as well as PhD students Jianming ZENG and Sen GUO also made important contributions to the study. The study was supported by Hanming SHEN, Chair Professor in the FHS, and Daming GAO, researcher in the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. The Genomics, Bioinformatics and Single Cell Analysis Core and the Animal Research Core in the FHS also provided tremendous support for the study. This project was supported by the Science and Technology Development Fund, Macao SAR (File no: 111/2017/A, 0011/2019/AKP, 0004/2021/AKP, 0048/2019/A1, 0112/2019/A2, 0065/2021/A and 0034/2019/AGJ), UM (File no: CPG2022-00002-FHS and MYRG2022-00181-FHS), and the National Natural Science Foundation of China (82030094 and 81602587). A complete version of the article can be viewed at https://www.science.org/doi/10.1126/sciadv.abq1395.