A team led by Ruiyu XIE, Associate Professor in the Faculty of Health Sciences (FHS) of the University of Macau (UM), has discovered the essential role of DNA demethylase TET1 in beta-cell specification. The discovery will help people understand TET family dioxygenases in cell fate determination, which has profound implications for beta-cell regeneration in cell based-therapies for diabetes. The study has been published in Nature Communications. It has received considerable attention in the field of pancreas research and has secured the No 1 spot of the 2022 Top Tweet Countdown in STEMCELL Tech’s Pancreatic Cell News.
Prof. Xie’s team previously demonstrated that 5-hydroxymethylcytosine (5hmC), a novel epigenetic modification, is positively correlated with ‘open’ chromatin at poised and active enhancers in multiple endodermal lineage intermediates. While many researchers had illustrated the importance of TET-regulated DNA demethylation homeostasis during early embryonic development and cell reprogramming, the functional relevance and mechanisms by which TETs regulate pancreatic endocrine differentiation remained obscure. For this reason, an in-depth understanding of how TETs function in beta-cell specification is important and will facilitate medical translational applications for beta-cell regeneration and reprogramming.
The research team set out to tackle this issue using a stepwise hESC differentiation system toward pancreatic endocrine fate, in which TET single-, double-, and triple-knockout hESCs were differentiated through multiple lineage intermediates into pancreatic endocrine cells. The team found that TET1/TET2/TET3 triple-knockout (TKO) hESCs could efficiently differentiate into definitive endoderm, but fail to generate functional beta-cells. Intriguingly, the team found extensive differentially methylated regions in a cell type-specific manner. In particular, the hyper-differential methylated regions specific to the pancreas displayed reduced chromatin activity and remarkable enrichment for the binding of FOXA2, a pioneer transcription factor essential for pancreatic endoderm specification. They further found that TET1 was crucial in the induction of functional beta-cell in vivo and demonstrated that FOXA2 favours TET1 deposition to facilitate local chromatin remodelling after pancreatic lineage induction.
The findings suggest that TET inactivation-induced aberrant chromatin remodelling at distinct FOXA2 binding sites contributes to defective beta-cell commitment and provides a new mechanistic clue regarding the complex crosstalk between epigenetic modifiers and pioneer transcription factors in lineage specification. The results of the exploration of how TET dioxygenases function in the pancreatic beta-cell specification can be applied to translational medicine for diabetes, such as the development of direct reprogramming of beta-cell. The study has secured the No 1 spot in the 2022 Top Tweet Countdown of STEMCEL Tech’s Pancreatic Cell News, which is a testament to its great impact and an indication of the broad reader interest.
Prof. Xie is the corresponding author of the study, and FHS PhD students Xinwei WU and Jie KE¸ as well as bioinformatics consultant Jianfang LI share the co-first authorship. Others who have contributed to the study include Prof. Deqiang SUN at Zhejiang University, who is the co-corresponding author of the study; Associate Professor Yun HUANG and her research team at Texas A&M University; as well as FHS former research assistant Qingping LAN and members of the Biological Imaging and Stem Cell Core and the Animal Research Core of FHS. The project was supported by the Science and Technology Development Fund, Macao SAR (File number: 0002/2021/AKP), and the National Natural Science Foundation of China (File number: NSFC 31701276). The full version of the research article can be viewed at https://www.nature.com/articles/s41467-022-31611-x.
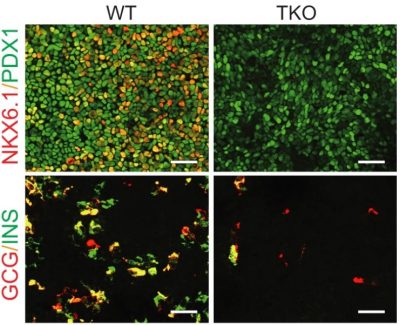
The immunostaining of PDX1/NKX6.1 and insulin/glucagon in wild-type and TET knockout cells
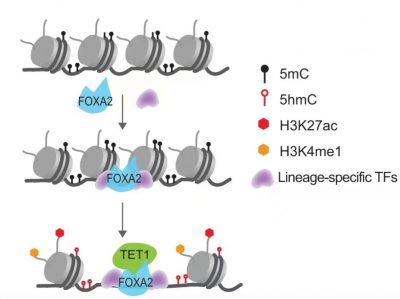
A schematic model depicting cooperative FOXA2 recruitment with lineage-specific transcription factors and TET1

Ruiyu XIE (3rd from right), Xinwei WU (4th from right), Jie KE (2nd from right) and Qingping LAN (1st from left)

